
I’m research scientist with expertise information theory, AI evaluation, and open-source scientific software. I did my PhD and postdoc in the UC Berkeley EECS department and Berkeley AI Research Lab, advised by Laura Waller.
My work includes:
- A technique to design cameras and other sensors for AI rather than for human vision
- The ARC-AGI-2 benchmark for testing fluid intelligence in large language models
- A 12-million-image biomedical dataset to train computer vision algorithms
- The most widely adopted software for automating microscopes, with applications ranging from cellular imaging to nanomaterials research
- Neural network architectures that incorporate physics knowledge to enable fast and low-cost scientific discovery
Email / Google scholar / GitHub / Bluesky
Research Highlights
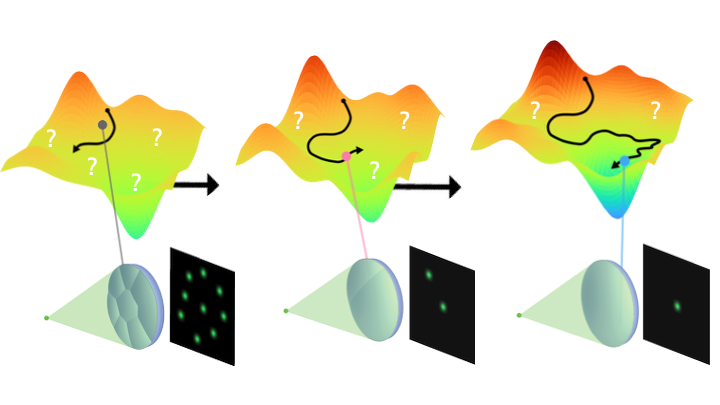
Information-driven design of imaging systems
NeurIPS 2025 website / paper / poster / code
A method to design imaging systems that capture maximum information for AI, free from human perceptual constraints. Applicable to diverse systems from consumer cameras to radio telescopes imaging black holes.
NeurIPS 2025 website / paper / poster / code
A method to design imaging systems that capture maximum information for AI, free from human perceptual constraints. Applicable to diverse systems from consumer cameras to radio telescopes imaging black holes.
 ARC-AGI-2 - A new challenge for frontier AI reasoning systems
ARC-AGI-2 - A new challenge for frontier AI reasoning systems
2025 website / paper
A benchmark for testing fluid intelligence and compositional reasoning in AI systems. Tests symbolic interpretation, multi-step reasoning, and contextual understanding beyond pattern matching.
The missing data for intelligent scientific instruments
Nature Methods 2025 paper
Outlines a path for AI to learn expert experimental skills from the rich digital traces that scientific instruments already generate.
Nature Methods 2025 paper
Outlines a path for AI to learn expert experimental skills from the rich digital traces that scientific instruments already generate.
The Berkeley single-cell computational microscopy (BSCCM) dataset
2024 website / paper / code
A 12-million-image biomedical computer vision benchmark with images of white blood cells under varied illumination, paired with protein expression labels.
2024 website / paper / code
A 12-million-image biomedical computer vision benchmark with images of white blood cells under varied illumination, paired with protein expression labels.
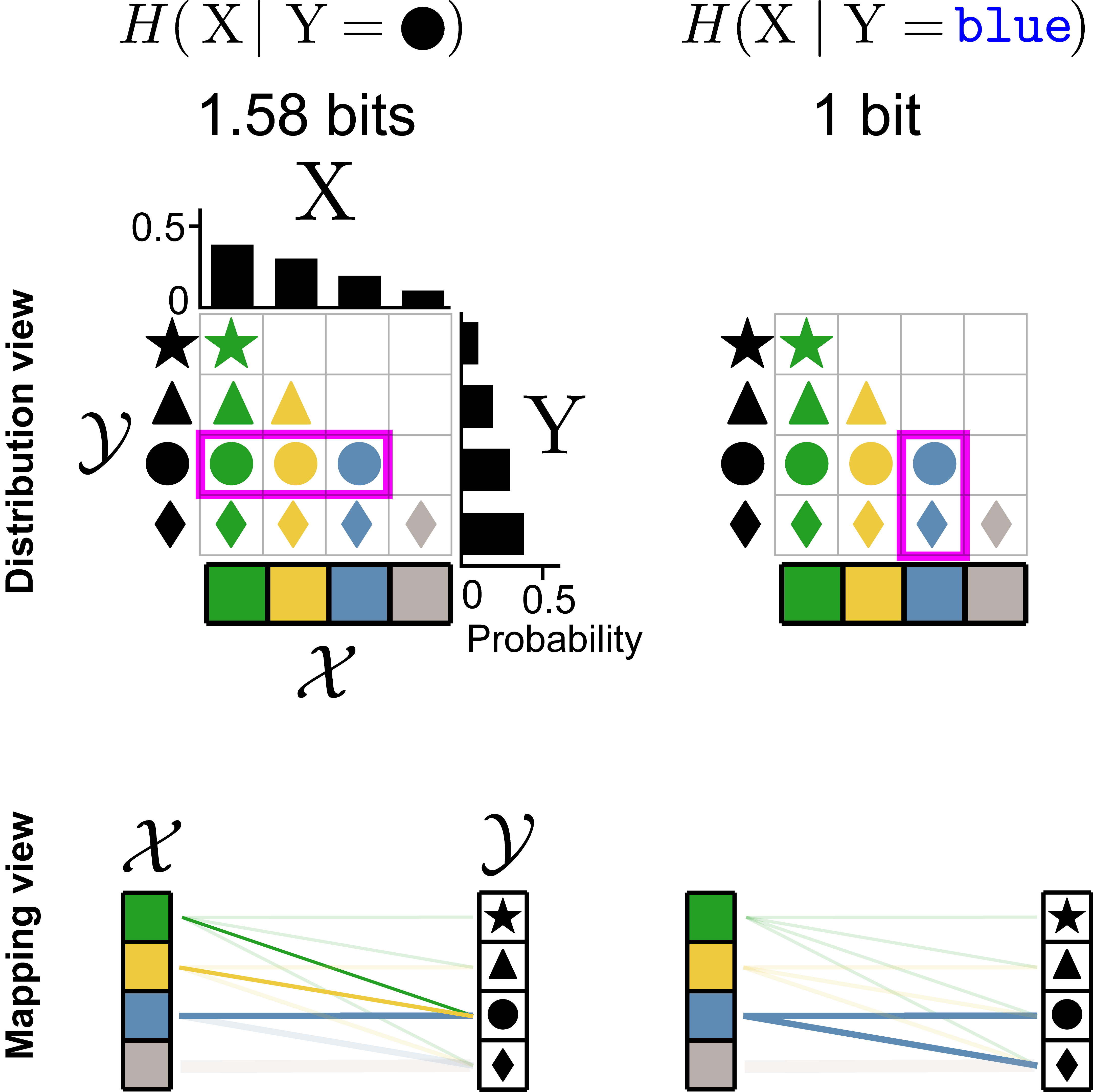 A visual introduction to information theory
A visual introduction to information theory
2022 website / paper / code+figures
Tutorial introduction to information theory, covering data compression and transmission in noisy channels.
 Pycro-Manager: open-source microscope control software
Pycro-Manager: open-source microscope control software
Nature Methods 2021 documentation / paper / code
Open-source Python package for microscope control, enabling automated experiments and real-time adaptive imaging. Works with hundreds of hardware components and handles terabyte-scale data acquisition.
Learned adaptive multiphoton illumination microscopy
Nature Communications 2021 paper / tutorial / data
Physics-informed neural networks that learn to compensate for optical scattering by dynamically adjusting laser power during scanning, enabling immune cell imaging at previously impossible depths in living tissue.
Nature Communications 2021 paper / tutorial / data
Physics-informed neural networks that learn to compensate for optical scattering by dynamically adjusting laser power during scanning, enabling immune cell imaging at previously impossible depths in living tissue.
Deep learning for single-shot autofocus microscopy
Optica 2019 paper / tutorial / code
Physics-informed neural network architecture that predicts focus corrections from single images using custom illumination patterns, reducing parameters by 100× while maintaining accuracy.
Optica 2019 paper / tutorial / code
Physics-informed neural network architecture that predicts focus corrections from single images using custom illumination patterns, reducing parameters by 100× while maintaining accuracy.
Additional Publications
 Information-theoretic Bayesian optimization of imaging systems
Information-theoretic Bayesian optimization of imaging systems
Optica COSI 2025 website
Black-box framework for designing imaging systems using Bayesian optimization and mutual information, requiring neither forward models nor ground truth data.
 Computationally efficient information-driven optical design with interchanging optimization
Computationally efficient information-driven optical design with interchanging optimization
2025 paper
An application-agnostic optical design method with reduced runtime and memory for information-theoretic optimization of imaging systems.
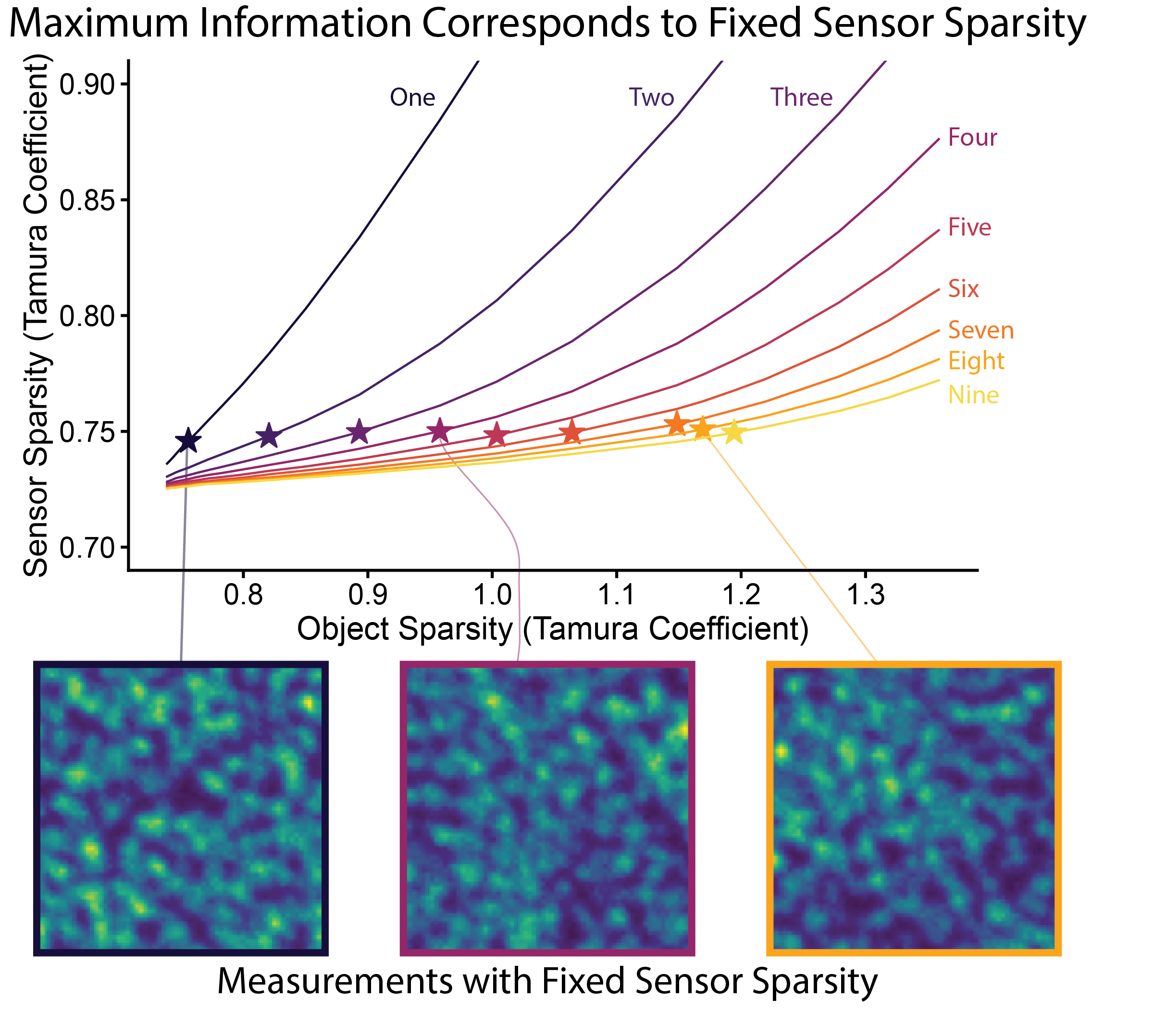 Designing lensless imaging systems to maximize information capture
Designing lensless imaging systems to maximize information capture
Optica 2026 website / paper / code
Mutual information-based evaluation and design of lensless imaging systems, establishing principles for object-dependent imaging.
 Neural network-based dark-field differential phase contrast microscopy
Neural network-based dark-field differential phase contrast microscopy
SPIE BiOS 2025 paper
Fast quantitative phase imaging method achieving high resolution with fewer images than traditional approaches.
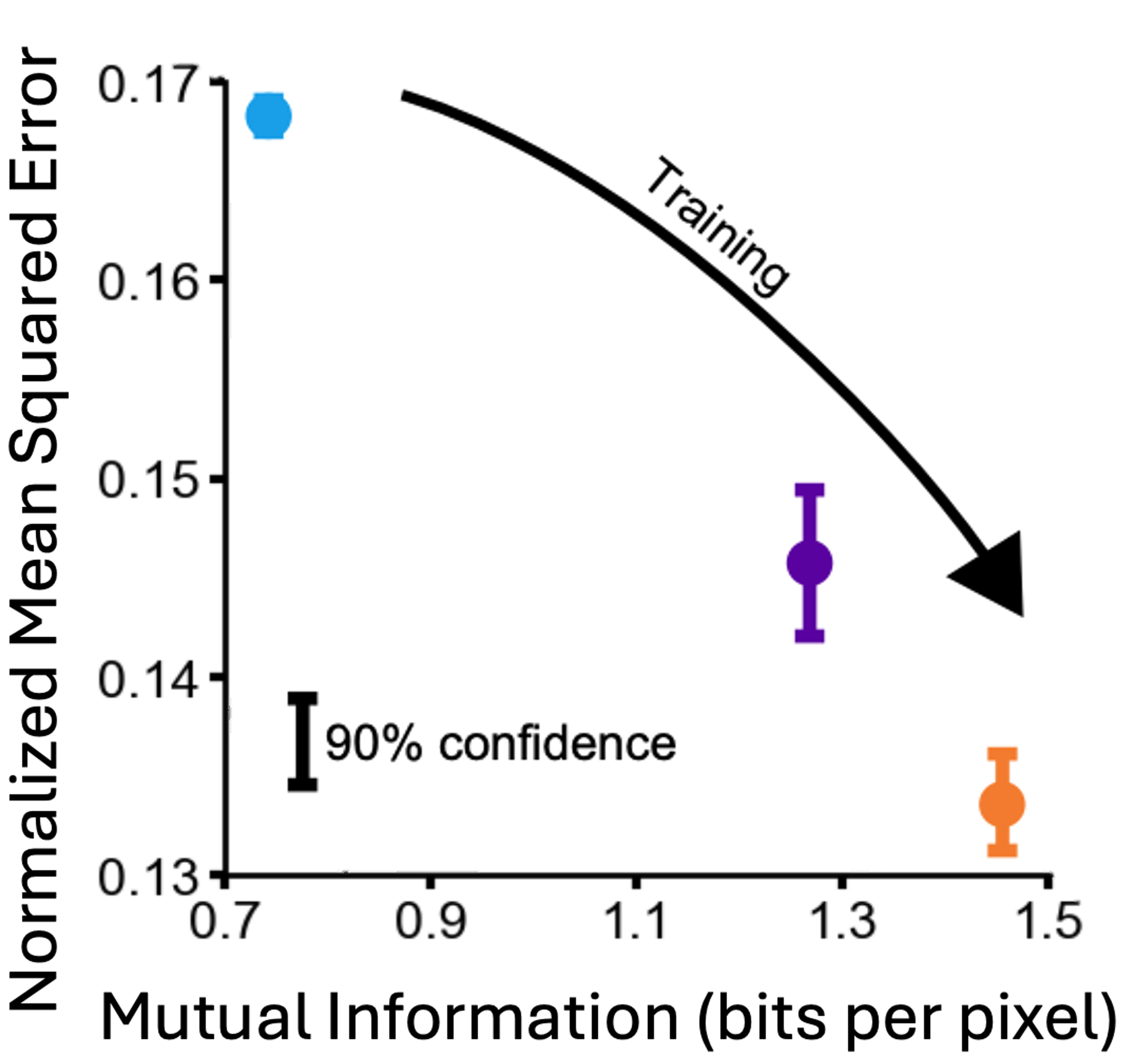 Information-theoretic design for high-dimensional computational imaging
Information-theoretic design for high-dimensional computational imaging
Optica COSI 2024
A technique for information-theoretic optimization of computational imaging systems with high-dimensional design spaces.
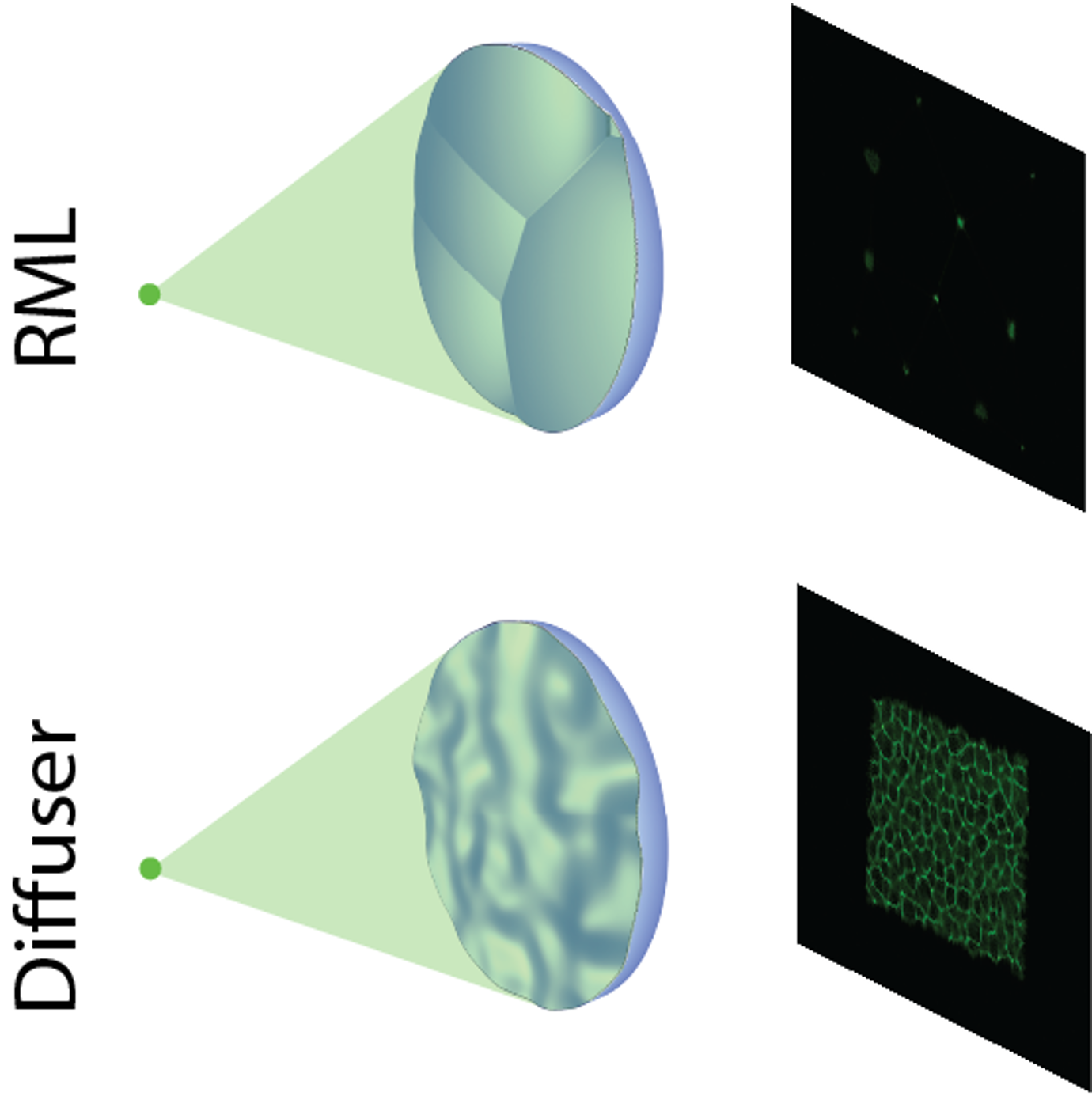 Information-theoretic experimental analysis of lensless imagers
Information-theoretic experimental analysis of lensless imagers
Optica COSI 2024
Analysis of information content in experimental lensless cameras showing that mutual information predicts reconstruction quality without performing reconstruction.
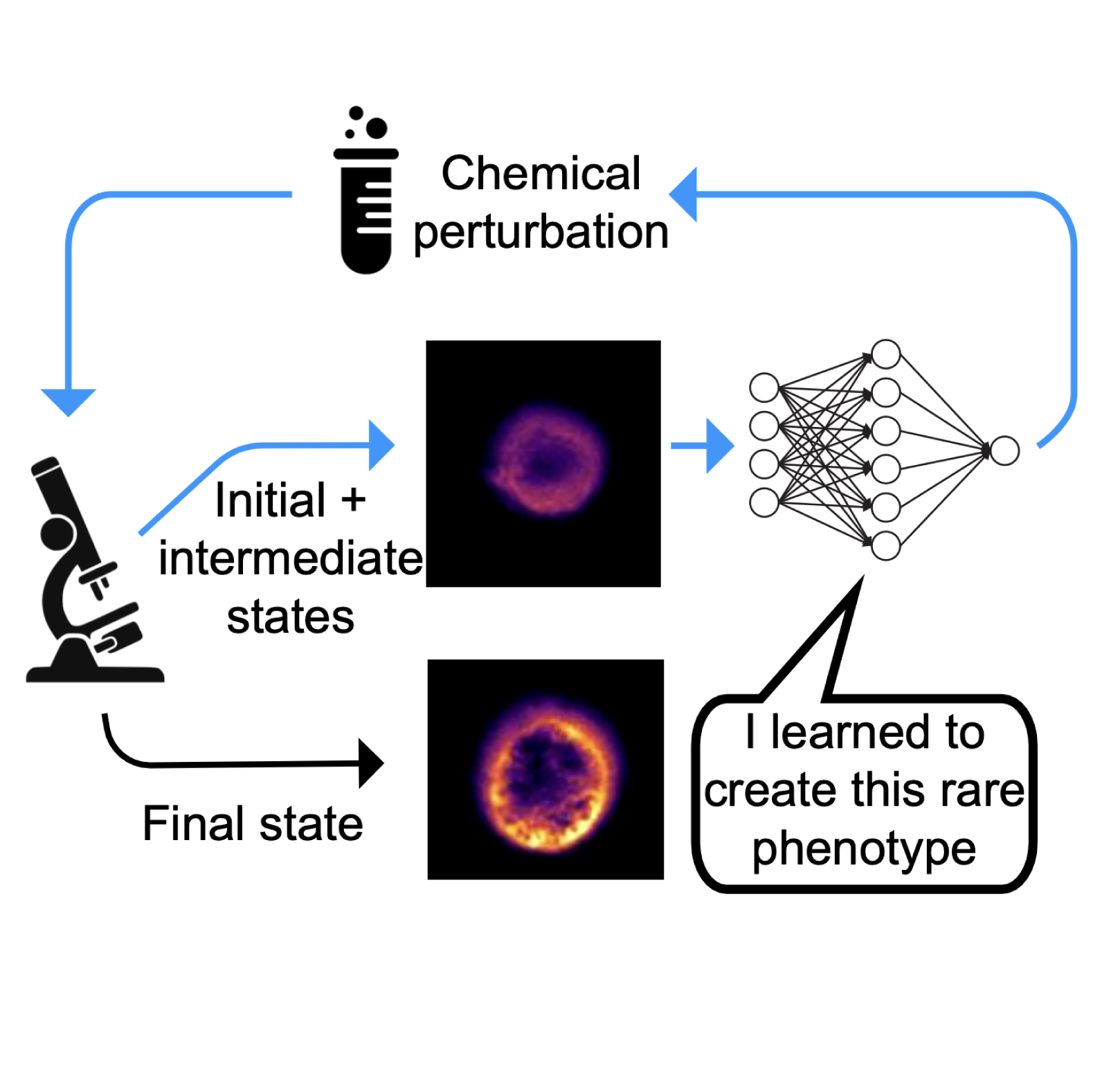 Microscopes are coming for your job
Microscopes are coming for your job
Nature Methods 2022 paper
Perspective on future possibilities for agentic AI and reinforcement learning in automated scientific discovery.
 Image denoising for fluorescence microscopy by supervised to self-supervised transfer learning
Image denoising for fluorescence microscopy by supervised to self-supervised transfer learning
Optics Express 2021 paper
Deep learning method for removing noise from microscopy images without requiring matched training data.
 The Microscope Device Abstraction Layer of Micro-Manager
The Microscope Device Abstraction Layer of Micro-Manager
2021 paper
Software architecture enabling control of thousands of microscope devices through a unified interface.
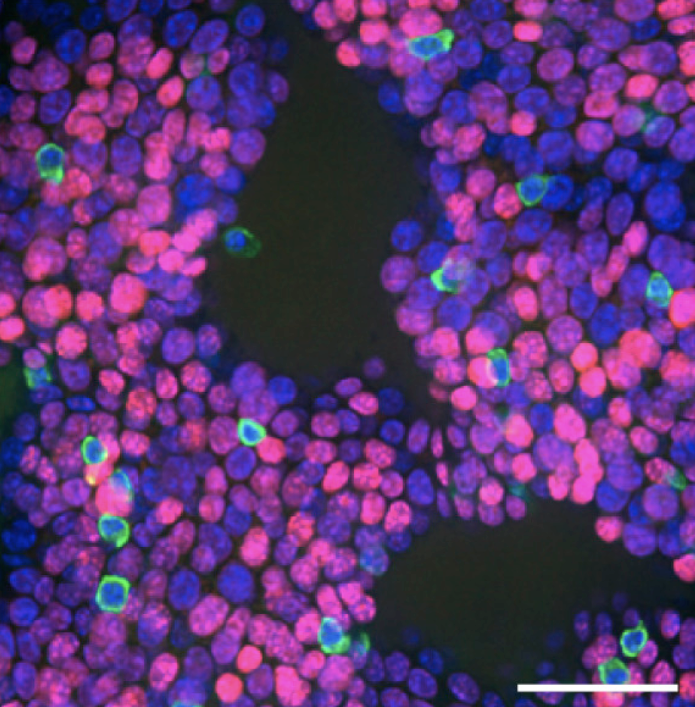 Quantitative Clonal Analysis and Single-Cell Transcriptomics Reveal Division Kinetics, Hierarchy, and Fate of Oral Epithelial Progenitor Cells
Quantitative Clonal Analysis and Single-Cell Transcriptomics Reveal Division Kinetics, Hierarchy, and Fate of Oral Epithelial Progenitor Cells
Cell Stem Cell 2019 paper
Single-cell analysis revealing the organization and dynamics of oral tissue stem cells.
Micro-Magellan
Nature Methods 2016 documentation / paper / code
Software for automated 3D imaging of large biological samples.
Nature Methods 2016 documentation / paper / code
Software for automated 3D imaging of large biological samples.
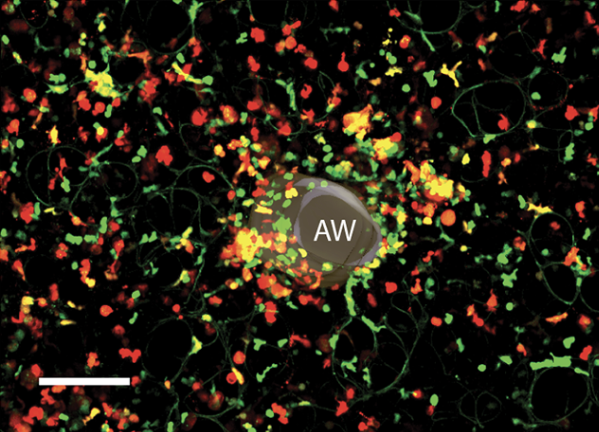 Tracking the Spatial and Functional Gradient of Monocyte-To-Macrophage Differentiation in Inflamed Lung
Tracking the Spatial and Functional Gradient of Monocyte-To-Macrophage Differentiation in Inflamed Lung
PLOS ONE 2016 paper
Imaging technique for tracking immune cell development in inflamed tissue.
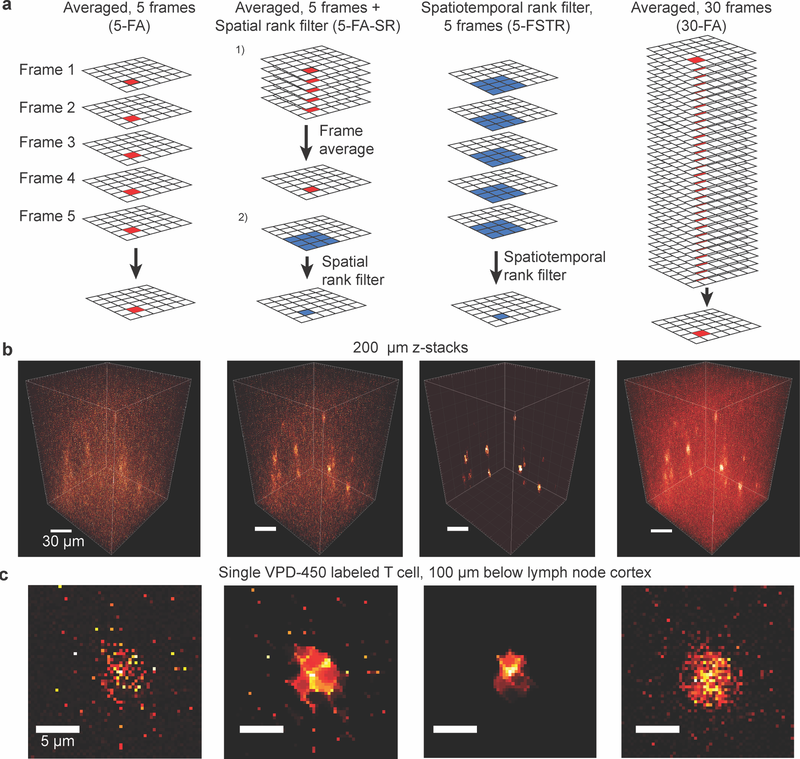 Spatiotemporal Rank Filtering Improves Image Quality Compared to Frame Averaging in 2-Photon Laser Scanning Microscopy
Spatiotemporal Rank Filtering Improves Image Quality Compared to Frame Averaging in 2-Photon Laser Scanning Microscopy
PLOS ONE 2016 paper
Computational method for improving microscope image quality while reducing exposure time.
 Assessing and benchmarking multiphoton microscopes for biologists
Assessing and benchmarking multiphoton microscopes for biologists
Methods in Cell Biology 2014 paper
Guidelines for evaluating and optimizing multiphoton microscope performance for biological imaging.
 Advanced methods of microscope control using μManager software
Advanced methods of microscope control using μManager software
Journal of Biological Methods 2014 paper
Open-source software platform for controlling automated microscopes and coordinating complex imaging experiments.
 Limb phase flexibility in walking: a test case in the squirrel monkey (Saimiri sciureus)
Limb phase flexibility in walking: a test case in the squirrel monkey (Saimiri sciureus)
Frontiers in Zoology 2019 paper
Model for predicting quadrupedal gait selection based on energy efficiency and balance requirements.
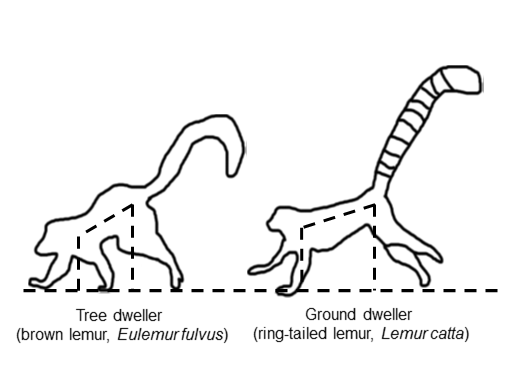 Pitch control and speed limitation during overground deceleration in lemurid primates
Pitch control and speed limitation during overground deceleration in lemurid primates
Journal of Morphology 2019 paper
Study showing how primate body geometry helps avoid forward pitching during rapid deceleration.
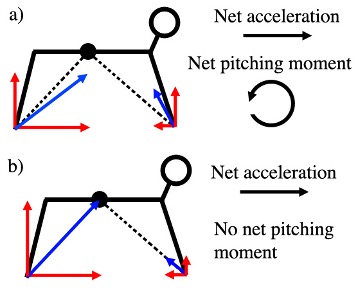 The mechanics of acceleration and deceleration in primate quadrupeds
The mechanics of acceleration and deceleration in primate quadrupeds
FASEB Journal 2013 paper
Analysis of how primate anatomy affects their ability to accelerate and decelerate during movement.